One goal of GLIMPRINT is to create web-based resource where models relevant to immunology and digital twins can be hosted. Easy access to models in a runnable form is critical for education and for communicating the capabilities (and limitations) of computational models. The model should include a clear description of what is included, and should allow the user to run the model easily with the capability of modifying parameters and exploring outputs. We are particularly interested in the educational power of basic models that can be run interactively in a web browser.
Models will cover a range of complexities, from simple viral infection SIR (susceptible - infected - recovered) models in JavaScript or SBML running in a browser or Jupyter Notebook (or as a NetLogo model), to complex multiscale models that include discreet cells, subcellular processes, immune response, etc.
We list here three types of models: (1) Ones that can be run directly here on GLIMPRINT web pages, (2) ones the can be run in a web browser at other online sites, and (3) models that require download of the model and needed software.
Models that can be directly run on GLIMPRINT web pages.
- A simple infection SIR (susceptible - infected - recovered) model in JavaScript that will run directly in your browser.
This model was created by Lorenzo Felletti whom we thank for sharing.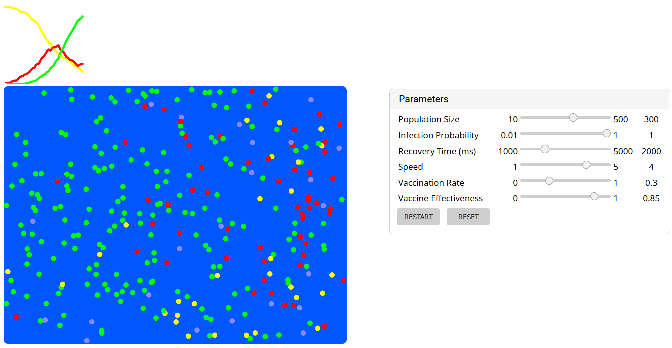
Models that can be run in a web browser at a site other then GLIMPRINT.
- Models hosted at nanoHUB
nanoHUB is funded by the NSF and others, and is an online computing host for a range of computational models in nanotechnology, physics and biology. NanoHUB supports a wide range of computing languages and has been used to tech both scientific and computing techniques. Running a model requires a user account, which is free. See https://nanohub.org/, to register visit https://nanohub.org/register/. Note that you must be a registered user and logged in to nanoHUB with your browser before the links to models given below will work properly. - Sego et al: Multiscale, multicellular, spatiotemporal modeling of acute primary viral infection and immune response in epithelial tissues. (Click the link to go to the GLIMPRINT page with more info.)
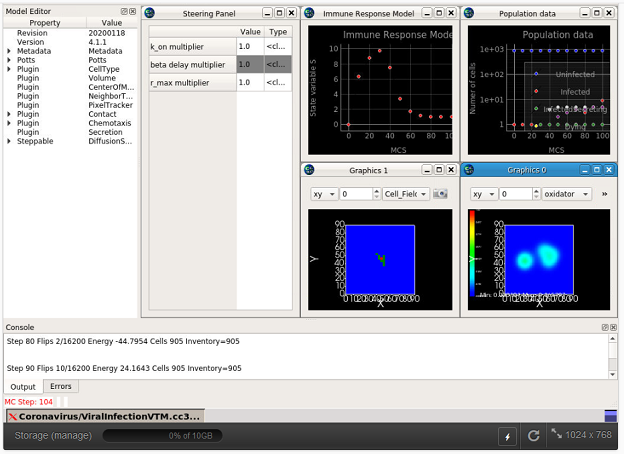
- Models runnable on NetLogo
NetLogo (https://ccl.northwestern.edu/netlogo is is a modeling language for agent-based models (ABMs). It can be run as either a local application on your computer or remotely via a web browser. - Cockrell and An, Comparative Computational Modeling of the Bat and Human Immune Response to Viral Infection with the Comparative Biology Immune Agent Based Model (Click the link to go to the GLIMPRINT info page.)
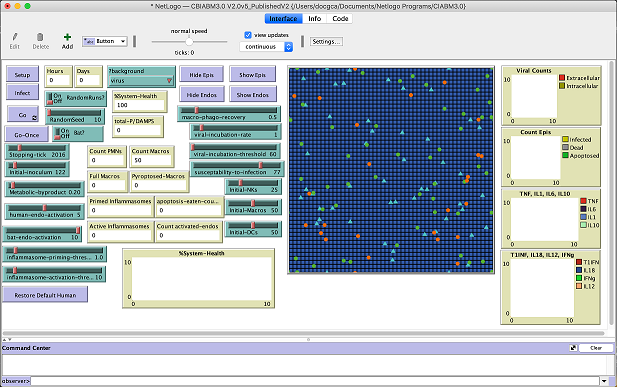
Models that must be downloaded along with the suitable program to run the model. We will limit to only software packages that are free and open source.
- tbd